OMOP Clinical Data Structure
Overview
The clinical data in AI-READI is formatted according to the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM), a commonly used framework for representing health data. OMOP has been used in many other well-known sources of data, such as the All of Us Research Program, the National COVID Cohort Collaborative (N3C), and the MIMIC database. All of Us has some overview information about OMOP here.
OMOP enables harmonization of data from multiple sources, for example enabling data from different electronic health record (EHR) systems to be formatted in the same way. This standard specifies both the structure and the content of the data. We have used OMOP to represent clinical data in AI-READI, including clinical lab tests, cognitive testing results (MoCA), monofilament testing, physical assessment, vision assessment, and survey questionnaires. We have written this guide to help orient users of AI-READI who may not have had prior experience working with OMOP-formatted data. Much more detailed information can be found on the OHDSI website and in the Book of OHDSI, which were the sources of much of the material described below. Additional guidance is also available at the OMOP Common Data Model GitHub page.
Background regarding the OMOP Common Data Model
The OMOP Common Data Model has standardized tables to represent health data, and it leverages an array of well-known standardized vocabularies, including the International Classification of Disease (ICD), the Systematized Nomenclature of Medicine (SNOMED), Logical Observation Identifiers Names and Codes (LOINC), RxNorm, and more. The figure below (from www.ohdsi.org) provides a schematic overview of the data model:
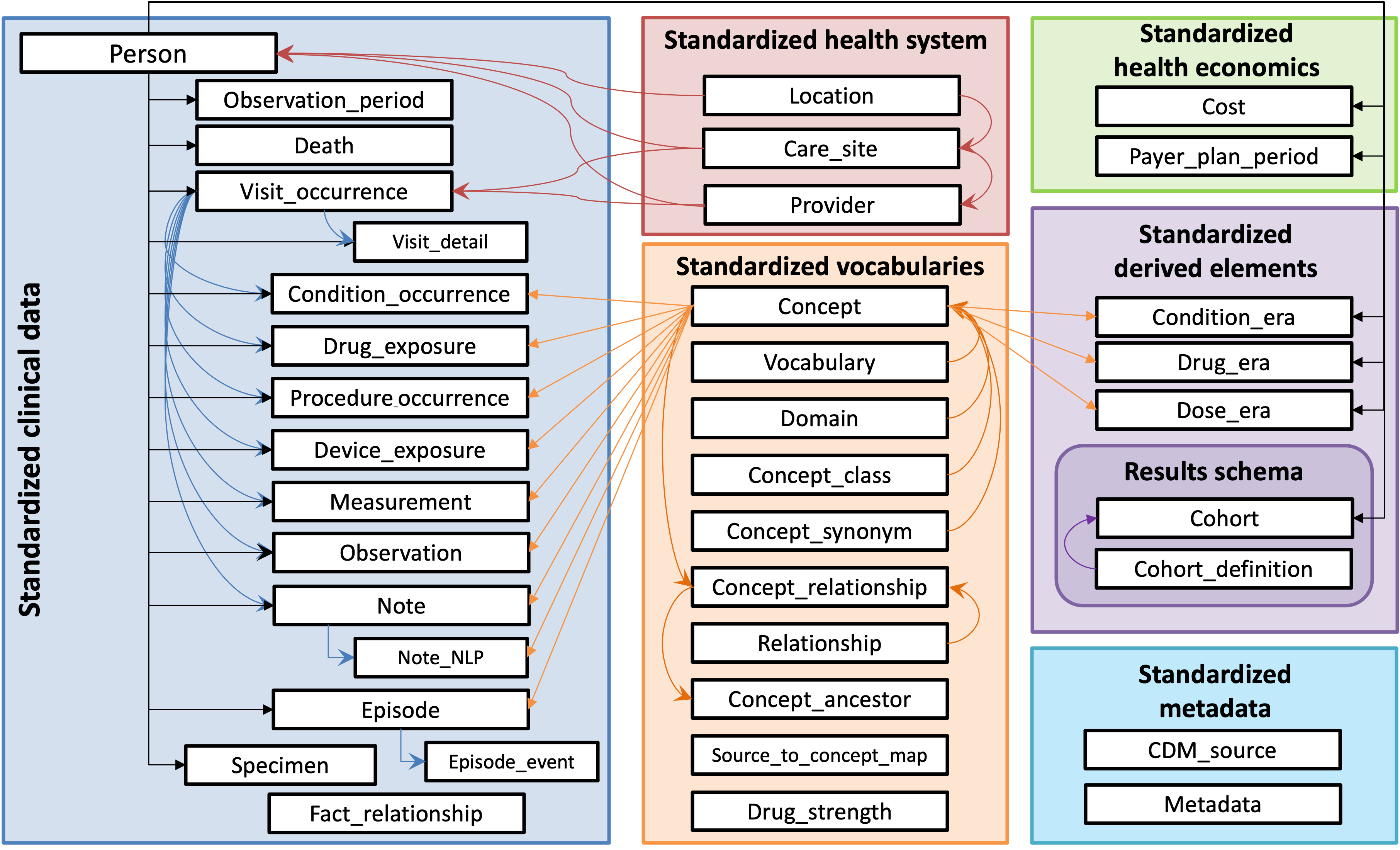
This data model is person-centric. Therefore, all of the clinical data tables are linked to the PERSON table. When you are working with AI-READI data, each person_id represents a unique participant, and the clinical data encompassed across the various tables can be linked by person_id, which is a column that is located in each table (i.e. it serves as a foreign key across the dataset).
Naming Conventions
Concepts are central to the OMOP Common Data Model. Concepts are standardized representations of each data element. So for example, if a data source has a particular patient where 'type 2 diabetes' is listed in the medical history, then in OMOP format, the standard concept would be the corresponding OMOP concept ID and it would be listed in the Conditions table, and this OMOP concept ID would harmonize/subsume other representations such as ICD or SNOMED codes. Each of the standard tables represents a domain of OMOP (e.g. procedures, conditions, measurements, drug exposures, etc.). The tables corresponding to each domain have the naming convention below (original source: Chapter 4 in the Book of OHDSI):
| Notation | Description |
|---|---|
| [Domain]_ID | Unique identifier for each record, which serves as a foreign key establishing relationships across Domain tables. For example, PERSON_ID uniquely identifies each individual. VISIT_OCCURRENCE_ID uniquely identifies a Visit. |
| [Domain]_CONCEPT_ID | Foreign key to a Standard Concept record in the CONCEPT reference table. This is the main representation of the Event, serving as the primary basis for all standardized analytics. For example, CONDITION_CONCEPT_ID = 31967 contains the reference value for the SNOMED concept of 'Nausea'. |
| [Domain]_SOURCE _CONCEPT_ID | Foreign key to a record in the CONCEPT reference table. This Concept is the equivalent of the Source Value (below), and it may happen to be a Standard Concept, at which point it would be identical to the [Domain]_CONCEPT_ID, or another non-standard concept. For example, CONDITION_SOURCE_CONCEPT_ID = 45431665 denotes the concept of 'Nausea' in the Read terminology, and the analogous CONDITION_CONCEPT_ID is the Standard SNOMED-CT Concept 31967. The use of Source Concepts for standard analytics applications is discouraged since only Standard Concepts represent the semantic content of an Event in a unambiguous way and therefore Source Concepts are not interoperable. |
| [Domain]_TYPE_CONCEPT_ID | Foreign key to a record in the CONCEPT reference table, representing the origin of the source information, standardized within the Standardized Vocabularies. Note that despite the field name this is not a type of an Event, or type of a Concept, but declares the capture mechanism that created this record. For example, DRUG_TYPE_CONCEPT_ID discriminates if a Drug record was derived from a dispensing Event in the pharmacy ('Pharmacy dispensing') or from an e-prescribing application ('Prescription written') |
| [Domain]_SOURCE_VALUE | Verbatim code or free text string reflecting how this clinical data was represented in the source data. Its use is discouraged for standard analytics applications, as these Source Values are not harmonized across data sources. For example, CONDITION_SOURCE_VALUE might contain a record of ICD-10 CM code R11.0 that corresponds to 45534429 concept in OMOP concept vocabulary table. |
Examples in AI-READI Documentation
You will see in our documentation site that we have described these notations in detail for particular types of clinical data, including clinical lab tests, cognitive testing, monofilament testing, physical assessment, and vision assessment. By going to each of those individual pages, you can see which table contains that particular data type, what each column header is, a description for each, and example output in the data file.
For example, let's use the physical assessment information as a specific illustration. Physical assessment in AI-READI includes height, weight, body mass index, waist circumference, hip circumference, waist-to-hip ratio, heart rate, and blood pressure. By looking at the documentation page, you can see that if you are interested in analyzing these values from the dataset, you would find them in the 'measurement.csv' file in the Clinical Data folder. The final section on 'Metadata and Example Outputs' will walk you through how to interpret the file, and what each column in the header row represents. Note that there are multiple columns to help you identify the concept. There is a column listing the standard OMOP concept ID number (measurement_concept_id, e.g. '4245997') and a text description (measurement_source_value, e.g. 'BMI'). The reason the standard OMOP concept ID number is important is because different data sources can have different ways of representing 'BMI' - for example, one health system may represent it as 'BMI' while another represents it as 'Body Mass Index.' By mapping the various source descriptions to a common, standardized data element (concept ID 4245997), this allows for non-ambiguous representation and ability to then aggregate those data from different sources into a single data repository. In this particular study, we have a uniform protocol across all three data collection sites, so this is less of a harmonization issue at the level of the study itself (i.e., difficulties of harmonizing data across the 3 sites is reduced because we are starting out with a consistent method of data acquisition). However, this is important for downstream analyses wherein data from AI-READI may be used as a validation dataset for analyses performed using other datasets (or vice versa). Having data mapped to standardized terminologies and to a common data model facilitates reproducibility.
Table 2 provides an overview of each data type and which table associated variables can be found. This is a high-level overview guide so that if you are looking for a particular variable, you know which file to access to get information on that variable.
| Data Category | Variables | OMOP Table / CSV file within the AI-READI Clinical Data folder |
|---|---|---|
| Clinical Lab Tests | NT-proBNP, Troponin-T, C-peptid, Insulin, CRP-HS, Total Cholesterol, Triglycerides, HDL Cholesterol, LDL Cholesterol, Glucose, BUN, Creatinine, BUN/Creatinine ratio, Sodium, Potassium, Chloride, Total Carbon Dioxide, Calcium, Albumin, Total Globulin, A/G Ratio, Total Bilirubin, Alkaline Phosphatase, AST, ALT, HbA1c, Urine Creatinine, Urine Albumin | Measurement |
| Cognition | MoCA test output | Measurement |
| Monofilament Testing | Number of correct responses and number of sites tested for each foot | Measurement |
| Physical Assessment | Height, Weight, Body Mass Index, Waist Circumference, Hip Circumference, Waist-to-Hip Ratio, Heart Rate, and Blood Pressure | Measurement |
| Questionnaires | General categories include demographics, general health, diabetes self care, depression, social determinants of health, diet, substance use, and vision and access to eye care. For individual survey items, see the Questionnaires page for more information. Note, in OHDSI OMOP CDM, data for patients with negative events isn't typically recorded. However, to capture both negative and positive responses to survey questions, we're recording negative responses alongside positive ones in the observation domain, using the specific question ID as the observation_concept_id. | Most survey questions are mapped to the Observation domain. Some questions (for example, summary scores) are mapped to the Measurement domain. If a participant reports having a health condition/ clinical disorder, then the self reported clinical disorder or the condition is mapped to the proper condition_concept_id and is recorded in the Condition domain. |
| Vision Assessment | Autorefraction, Best Corrected Visual Acuity | Measurement |
| Mars Letter Contrast Sensitivity | Measurement |
Navigating the Tables
OMOP tables are typically long in format. For example, a given participant will have many different kinds of measurements. As a result, the 'measurement.csv' table will include all of the measurements for all of the participants, including the laboratory testing values, physical assessment measurements, and other types of testing. You will see person_ID numbers repeated as a result, as each row will include a separate measurement, and there will be numerous measurements per participant.
Some data users may be accustomed to wide tables, where each row represents a single participant, and each column represents a variable of interest, and the individual cells represent the data corresponding to that variable for that person. This format may be more amenable for use in downstream analyses, but it is unable to capture more complex health data where there are multiple values over time for a given participant, and is not compliant with the specified structure of the OMOP Common Data Model.
However, data users are certainly welcome to wrangle the AI-READI data from long format to wide format. There are many useful guides online about how to do this, and we will not replicate them here. But, we have provided some example code snippets in the individual documentation pages to assist with streamlining some of the tables if you would like to focus on the measurement values and don't need all of the other OMOP encodings for your downstream analyses.
Creating Cohorts
If you would like to use the entire cohort of participants in AI-READI, you can use the Person table ('person.csv') to see a list of all person_id numbers that are currently available in the dataset. This serves as the foreign key for all of the other OMOP clinical data tables (Conditions, Measurements, Procedures, etc.).
If you would like to create a cohort based on a set of specific criteria, you can query the tables to filter/subset the group of participants that meet your criteria and define the cohort that way. For example, let's say that you are interested in studying individuals with obesity and would like to limit your analyses to those with BMI values greater than or equal to 30. Based on the guide we provided in Table 2 above, you would know that BMI values are in the Measurements table ('measurement.csv'). You would use that file and could access BMI values (either using the measurement_concept_id column for concept ID 4245997 or the measurement_source_value column for BMI), and then subset the data to include only those with BMI of 30 or greater (in the value_as_number column). With that subsetted data frame, you would have the person_id numbers corresponding to obese individuals in the dataset. You would then use that as your study cohort, and would use that array of person_id numbers to perform joins with other data of interest from the various tables to generate a data frame used for your subsequent analyses.
Defining cohorts based on concepts can be tricky if you don't know how to identify concept IDs. In the subsequent sections, we will provide some resources as to how you can locate OMOP concept IDs that were used in AI-READI. These can be used not only to create the baseline study cohorts, but also to create concept sets that will then comprise outcomes and predictors/covariates that will be used in AI/ML models.
Identifying Concepts
To find the standard OMOP concept corresponding to a particular variable, one way to search this is through the free, publicly available Athena terminology browser, which is managed by an organization called Observational Health Data Sciences and Informatics (OHDSI, pronounced like 'Odyssey') which oversees and manages the OMOP Common Data Model. It includes a free text search browser where you can search by a narrative descriptor or by specific source vocabularies (e.g. ICD codes).
For more specific information regarding the concept IDs used for AI-READI, the following section describes the mapping process (taking source data elements and finding the closest match in the OMOP Common Data Model). Using this resource below, you will have more information regarding the concept IDs used in the dataset and can subsequently define cohorts and concept sets accordingly.
Metadata on AI-READI OMOP Mappings
Over 2000 concepts were mapped to the OMOP CDM for the AI-READI project, with metadata on the quality of the mappings provided. A document explaining the mappings and how each data element in the AI-READI dataset corresponds to OMOP concepts is provided in the table below.
A Simple Standard for Sharing Ontological Mappings (SSSOM) Simple Knowledge Organization System (SKOS) categorizations were provided to denote the relationship between each concept and mapping (e.g. exact, close, narrow, broad, related) as well as the confidence level of the mapping (0-100%). Providing such metadata on the quality of the mappings improves transparency on the quality of the mappings and helps ensure they adhere to FAIR data principles.
Concepts without OMOP CDM mappings available, or concepts with less precise mappings (e.g. with SSSOM SKOS categorizations of narrow, broad, or related and/or confidence level at 50% or lower) received temporary local code extensions and the project will advocate with OHDSI for the creation of new more precise OMOP CDM codes for those concepts. Over 500 concepts meet this criteria.
Resources to learn more about SSSOM:
- Overview - Simple Standard for Sharing Ontological Mappings (SSSOM)
- SSSOM - Which mapping predicate should I choose? Cheat sheet
- A Simple Standard for Sharing Ontological Mappings (SSSOM)
OMOP Mapping Table
See the table here
Race, ethnicity, gender, and date/month of birth
Note that the fields for race, ethnicity, gender, and date/month of birth are all coded as '0' in the person.csv file because this information is omitted from the data. Birthday has to be released with birth year only per the HIPAA law for Safe Harbor. Additionally, we have decided to remove race, ethnicity, and gender information from this public set of the AI-READI dataset to prevent stigmatizing findings. More details are available in the healthsheet regarding which variables were removed.
Additional resources
Consider starting with our example Jupyter notebooks to explore the dataset further.